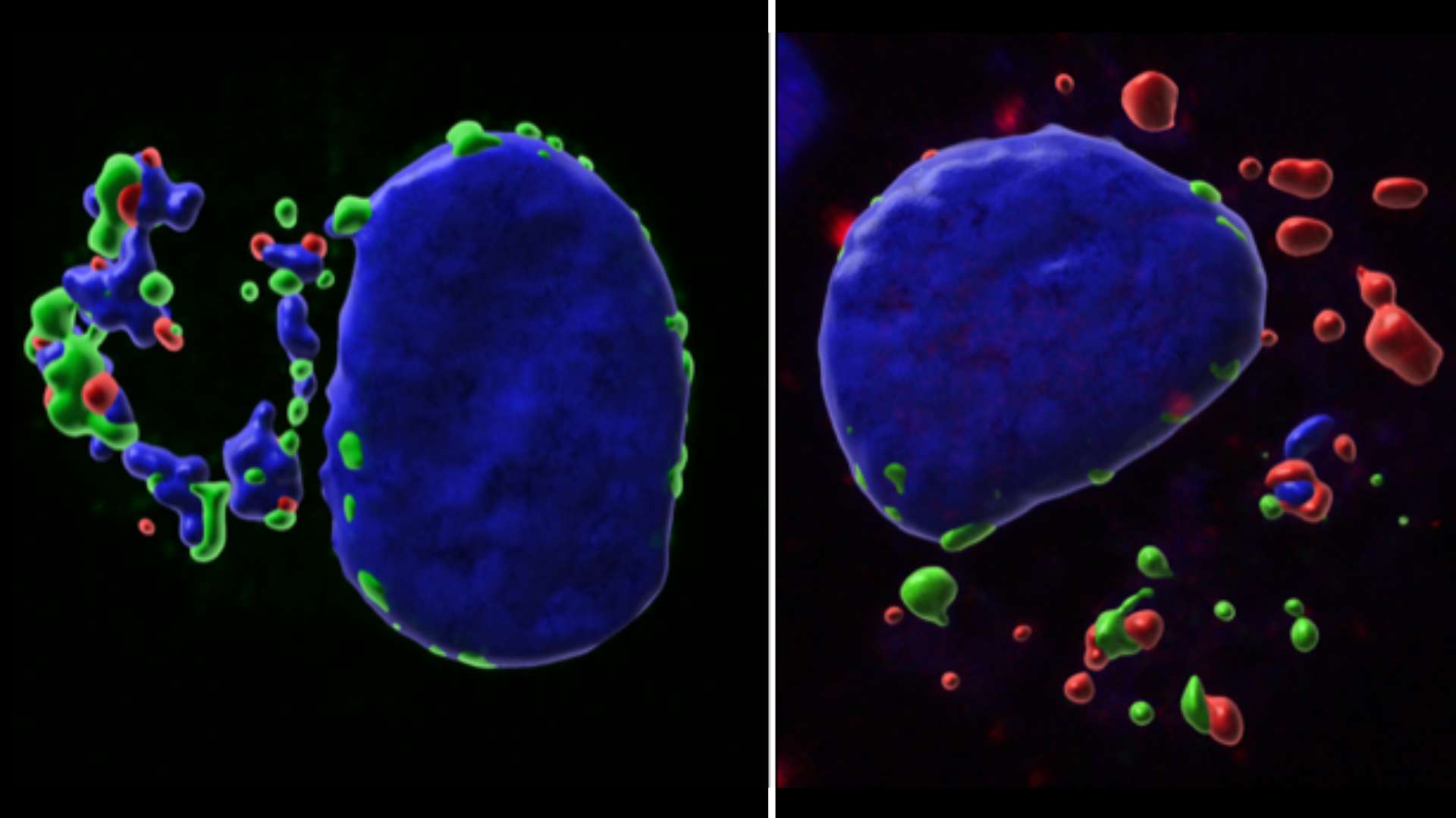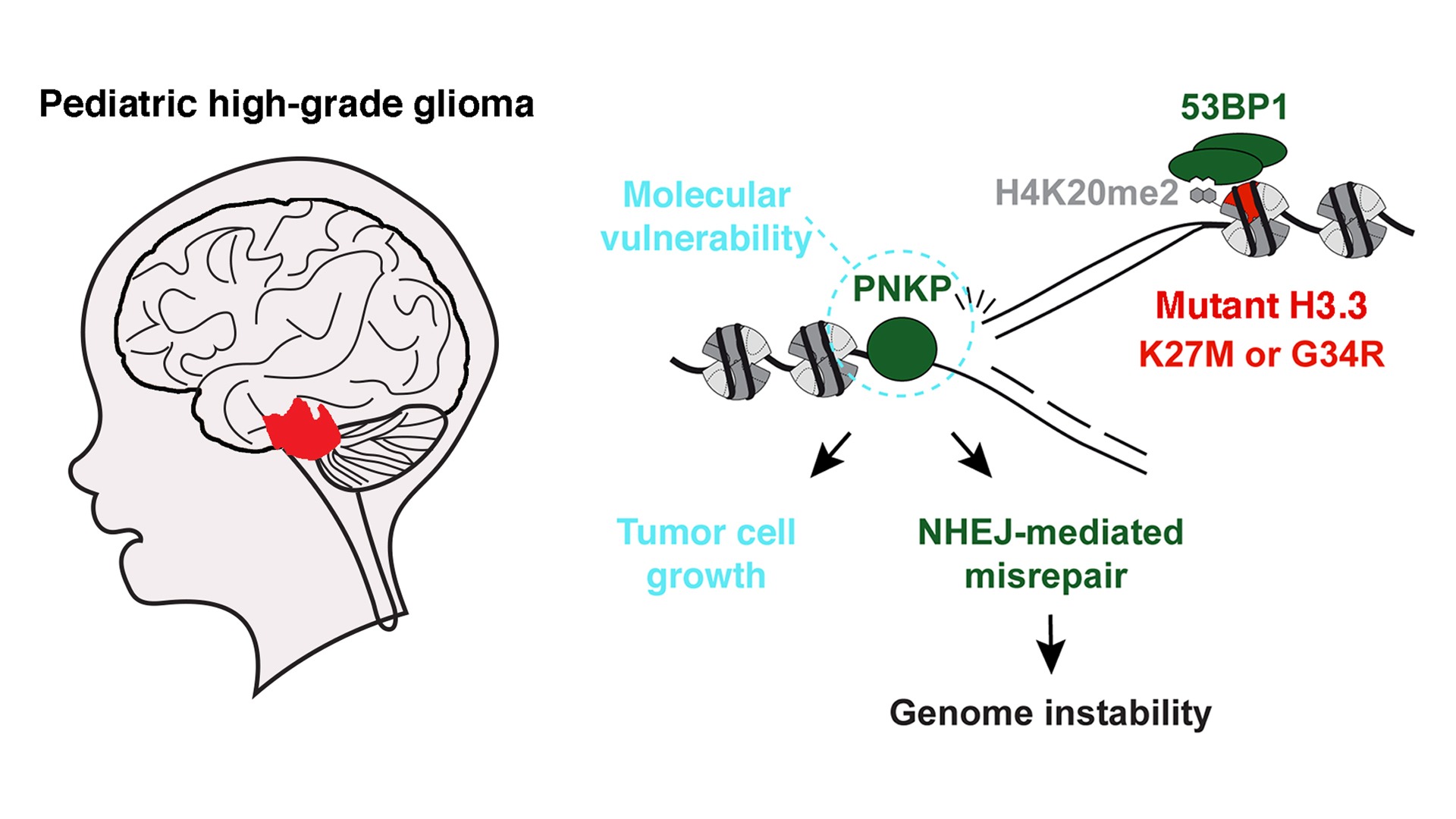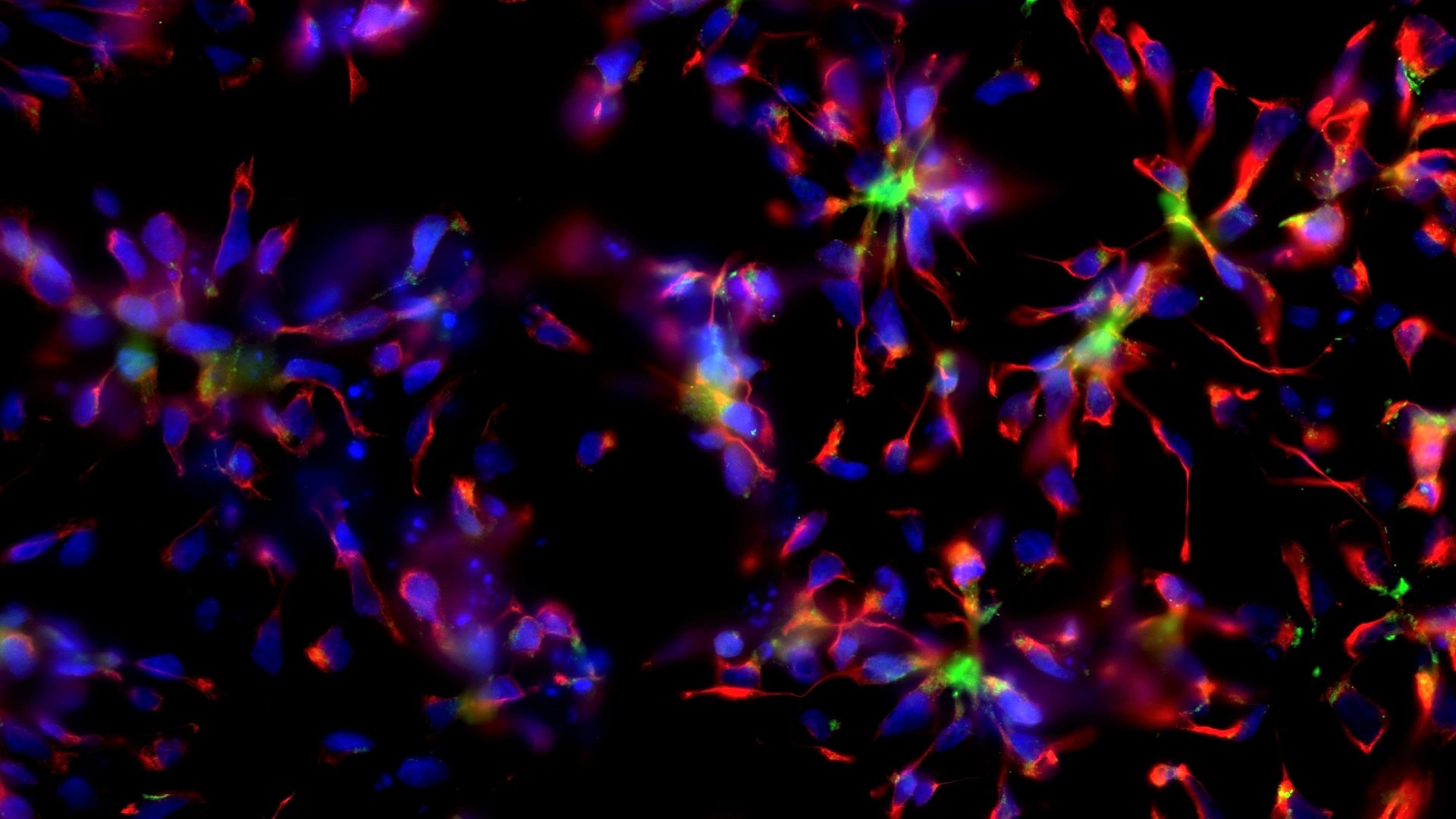
Epigenetics and Cell Fate Centre
Deciphering the epigenetic regulations determining cell fate and differentiation programs in mammals, under normal and pathological conditions
Focus

Our teams
Learn more about our six research teams.

Our platforms
Discover our platforms and technical facilities.
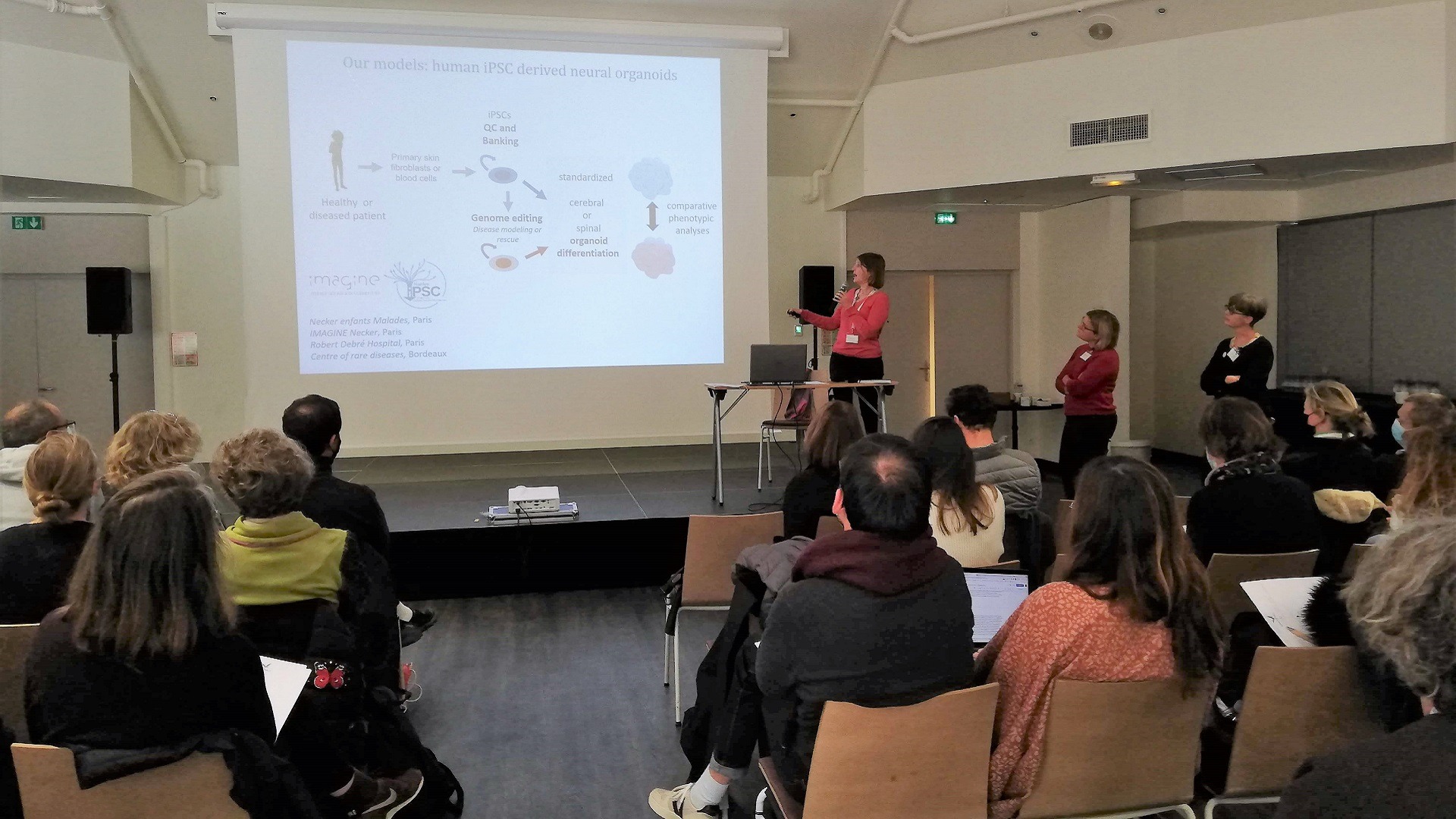
Transverse actions
Our centre contributes to transversal actions dedicated to training and research.

General public
Interact and communicate with the general public is part of our mission. Discover our actions.
News
Regulatory authorities